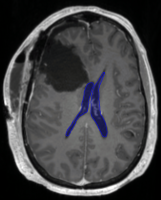
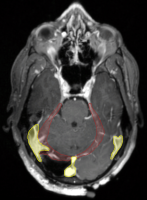
 The subject cohort includes patients diagnosed with glioblastoma and low-grade glioma who underwent surgery and radiotherapy treatment at Massachusetts General Hospital. As shown in the figure on the right, surgical cavities may deform and shift brain anatomy. The image data provided for training and validation, and testing consists of the CT scans acquired for treatment planning, and two diagnostic MRI scans, contrast enhanced T1-weighted and T2-weighted FLAIR, of the post-operative brain. The dataset includes images from 2 different CT scanners, and 7 different MRI scanners. All scans are available in MHA format: ct.mha, t1.mha, and t2.mha
The subject cohort includes patients diagnosed with glioblastoma and low-grade glioma who underwent surgery and radiotherapy treatment at Massachusetts General Hospital. As shown in the figure on the right, surgical cavities may deform and shift brain anatomy. The image data provided for training and validation, and testing consists of the CT scans acquired for treatment planning, and two diagnostic MRI scans, contrast enhanced T1-weighted and T2-weighted FLAIR, of the post-operative brain. The dataset includes images from 2 different CT scanners, and 7 different MRI scanners. All scans are available in MHA format: ct.mha, t1.mha, and t2.mha
All subject image sets were manually segmented to create a set of non-overlaying structures. For Task 1, the segmentation was done by one annotator and approved by a neuro-anatomist. For Task 2, multiple annotators, following the same annotation protocol, performed segmentation. The labels are available in MHA format separately for the two tasks.
The data was pre-processed: multi-modality images were co-registered, and re-sampled to the same resolution and size.
For training and validation, we will provide 45 multi-modal images and their ground truth annotations. There will be two testing stages. The first will include 15 multi-modal scans and will be used to provide participants with feedback through a challenge leaderboard. The second test stage will be a final test set (released close to the MICCAI meeting) of 15 multi-modal scans which will be used to for final scoring of participant’s algorithm performance. To remove possible batch effects stemming from annotator preferences or other factors, the 75 images are randomly assigned to sets of 45, 15, and 15 images.